I've been using cProfile to profile my code, and it's been working great. I also use gprof2dot.py to visualize the results (makes it a little clearer).
However, cProfile (and most other Python profilers I've seen so far) seem to only profile at the function-call level. This causes confusion when certain functions are called from different places - I have no idea if call #1 or call 开发者_高级运维#2 is taking up the majority of the time. This gets even worse when the function in question is six levels deep, called from seven other places.
How do I get a line-by-line profiling?
Instead of this:
function #12, total time: 2.0s
I'd like to see something like this:
function #12 (called from somefile.py:102) 0.5s
function #12 (called from main.py:12) 1.5s
cProfile does show how much of the total time "transfers" to the parent, but again this connection is lost when you have a bunch of layers and interconnected calls.
Ideally, I'd love to have a GUI that would parse through the data, then show me my source file with a total time given to each line. Something like this:
main.py:
a = 1 # 0.0s
result = func(a) # 0.4s
c = 1000 # 0.0s
result = func(c) # 5.0s
Then I'd be able to click on the second "func(c)" call to see what's taking up time in that call, separate from the "func(a)" call. Does that make sense?
I believe that's what Robert Kern's line_profiler is intended for. From the link:
File: pystone.py
Function: Proc2 at line 149
Total time: 0.606656 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
149 @profile
150 def Proc2(IntParIO):
151 50000 82003 1.6 13.5 IntLoc = IntParIO + 10
152 50000 63162 1.3 10.4 while 1:
153 50000 69065 1.4 11.4 if Char1Glob == 'A':
154 50000 66354 1.3 10.9 IntLoc = IntLoc - 1
155 50000 67263 1.3 11.1 IntParIO = IntLoc - IntGlob
156 50000 65494 1.3 10.8 EnumLoc = Ident1
157 50000 68001 1.4 11.2 if EnumLoc == Ident1:
158 50000 63739 1.3 10.5 break
159 50000 61575 1.2 10.1 return IntParIO
You could also use pprofile(pypi). If you want to profile the entire execution, it does not require source code modification. You can also profile a subset of a larger program in two ways:
toggle profiling when reaching a specific point in the code, such as:
import pprofile profiler = pprofile.Profile() with profiler: some_code # Process profile content: generate a cachegrind file and send it to user. # You can also write the result to the console: profiler.print_stats() # Or to a file: profiler.dump_stats("/tmp/profiler_stats.txt")toggle profiling asynchronously from call stack (requires a way to trigger this code in considered application, for example a signal handler or an available worker thread) by using statistical profiling:
import pprofile profiler = pprofile.StatisticalProfile() statistical_profiler_thread = pprofile.StatisticalThread( profiler=profiler, ) with statistical_profiler_thread: sleep(n) # Likewise, process profile content
Code annotation output format is much like line profiler:
$ pprofile --threads 0 demo/threads.py
Command line: ['demo/threads.py']
Total duration: 1.00573s
File: demo/threads.py
File duration: 1.00168s (99.60%)
Line #| Hits| Time| Time per hit| %|Source code
------+----------+-------------+-------------+-------+-----------
1| 2| 3.21865e-05| 1.60933e-05| 0.00%|import threading
2| 1| 5.96046e-06| 5.96046e-06| 0.00%|import time
3| 0| 0| 0| 0.00%|
4| 2| 1.5974e-05| 7.98702e-06| 0.00%|def func():
5| 1| 1.00111| 1.00111| 99.54%| time.sleep(1)
6| 0| 0| 0| 0.00%|
7| 2| 2.00272e-05| 1.00136e-05| 0.00%|def func2():
8| 1| 1.69277e-05| 1.69277e-05| 0.00%| pass
9| 0| 0| 0| 0.00%|
10| 1| 1.81198e-05| 1.81198e-05| 0.00%|t1 = threading.Thread(target=func)
(call)| 1| 0.000610828| 0.000610828| 0.06%|# /usr/lib/python2.7/threading.py:436 __init__
11| 1| 1.52588e-05| 1.52588e-05| 0.00%|t2 = threading.Thread(target=func)
(call)| 1| 0.000438929| 0.000438929| 0.04%|# /usr/lib/python2.7/threading.py:436 __init__
12| 1| 4.79221e-05| 4.79221e-05| 0.00%|t1.start()
(call)| 1| 0.000843048| 0.000843048| 0.08%|# /usr/lib/python2.7/threading.py:485 start
13| 1| 6.48499e-05| 6.48499e-05| 0.01%|t2.start()
(call)| 1| 0.00115609| 0.00115609| 0.11%|# /usr/lib/python2.7/threading.py:485 start
14| 1| 0.000205994| 0.000205994| 0.02%|(func(), func2())
(call)| 1| 1.00112| 1.00112| 99.54%|# demo/threads.py:4 func
(call)| 1| 3.09944e-05| 3.09944e-05| 0.00%|# demo/threads.py:7 func2
15| 1| 7.62939e-05| 7.62939e-05| 0.01%|t1.join()
(call)| 1| 0.000423908| 0.000423908| 0.04%|# /usr/lib/python2.7/threading.py:653 join
16| 1| 5.26905e-05| 5.26905e-05| 0.01%|t2.join()
(call)| 1| 0.000320196| 0.000320196| 0.03%|# /usr/lib/python2.7/threading.py:653 join
Note that because pprofile does not rely on code modification it can profile top-level module statements, allowing to profile program startup time (how long it takes to import modules, initialise globals, ...).
It can generate cachegrind-formatted output, so you can use kcachegrind to browse large results easily.
Disclosure: I am pprofile author.
Just to improve @Joe Kington 's above-mentioned answer.
For Python 3.x, use line_profiler:
Installation:
pip install line_profiler
Usage:
Suppose you have the program main.py and within it, functions fun_a() and fun_b() that you want to profile with respect to time; you will need to use the decorator @profile just before the function definitions. For e.g.,
@profile
def fun_a():
#do something
@profile
def fun_b():
#do something more
if __name__ == '__main__':
fun_a()
fun_b()
The program can be profiled by executing the shell command:
$ kernprof -l -v main.py
The arguments can be fetched using $ kernprof -h
Usage: kernprof [-s setupfile] [-o output_file_path] scriptfile [arg] ...
Options:
--version show program's version number and exit
-h, --help show this help message and exit
-l, --line-by-line Use the line-by-line profiler from the line_profiler
module instead of Profile. Implies --builtin.
-b, --builtin Put 'profile' in the builtins. Use 'profile.enable()'
and 'profile.disable()' in your code to turn it on and
off, or '@profile' to decorate a single function, or
'with profile:' to profile a single section of code.
-o OUTFILE, --outfile=OUTFILE
Save stats to <outfile>
-s SETUP, --setup=SETUP
Code to execute before the code to profile
-v, --view View the results of the profile in addition to saving
it.
The results will be printed on the console as:
Total time: 17.6699 s
File: main.py
Function: fun_a at line 5
Line # Hits Time Per Hit % Time Line Contents
==============================================================
5 @profile
6 def fun_a():
...
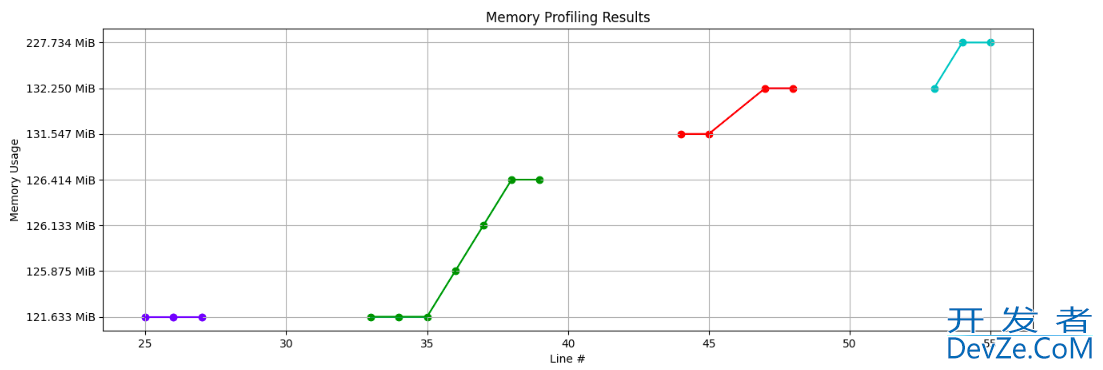
EDIT: The results from the profilers can be parsed using the TAMPPA package. Using it, we can get line-by-line desired plots as
You can take help of line_profiler package for this
1. 1st install the package:
pip install line_profiler
2. Use magic command to load the package to your python/notebook environment
%load_ext line_profiler
3. If you want to profile the codes for a function then
do as follows:
%lprun -f demo_func demo_func(arg1, arg2)
you will get a nice formatted output with all the details if you follow these steps :)
Line # Hits Time Per Hit % Time Line Contents
1 def demo_func(a,b):
2 1 248.0 248.0 64.8 print(a+b)
3 1 40.0 40.0 10.4 print(a)
4 1 94.0 94.0 24.5 print(a*b)
5 1 1.0 1.0 0.3 return a/b
PyVmMonitor has a live-view which can help you there (you can connect to a running program and get statistics from it).
See: http://www.pyvmmonitor.com/




![Interactive visualization of a graph in python [closed]](https://www.devze.com/res/2023/04-10/09/92d32fe8c0d22fb96bd6f6e8b7d1f457.gif)



 加载中,请稍侯......
加载中,请稍侯......
精彩评论